RNAs in the cell are highly decorated with proteins that control their function. We are particularly interested in understanding how the structure of an RNA affects the recognition by proteins and thereby ultimately impacts on cellular decisions. To identify novel functional RNA folds, we use the concept of evolutionary conservation.
The evolutionary conservation of sequences within an mRNA indicates their functional importance. The high conservation of protein coding sequences is particularly evident. Sequence conservation within the untranslated regions of an mRNA is usually low; but when it occurs, it signals important sites of post-transcriptional regulation. mRNAs, like other RNA molecules, are able to form complex three-dimensional structures. Thus, the evolutionary conservation of an mRNA fold is likewise indicative of a functional site.
Using bioinformatic prediction of mRNA folding and evolutionary conservation, we identified new binding sites for Roquin proteins. Roquin proteins recognize mRNAs shape-specifically and sequence-independently. The binding of Roquin to such so-called CDEs (constitutive decay elements) accelerates mRNA decay (Fig. 1).
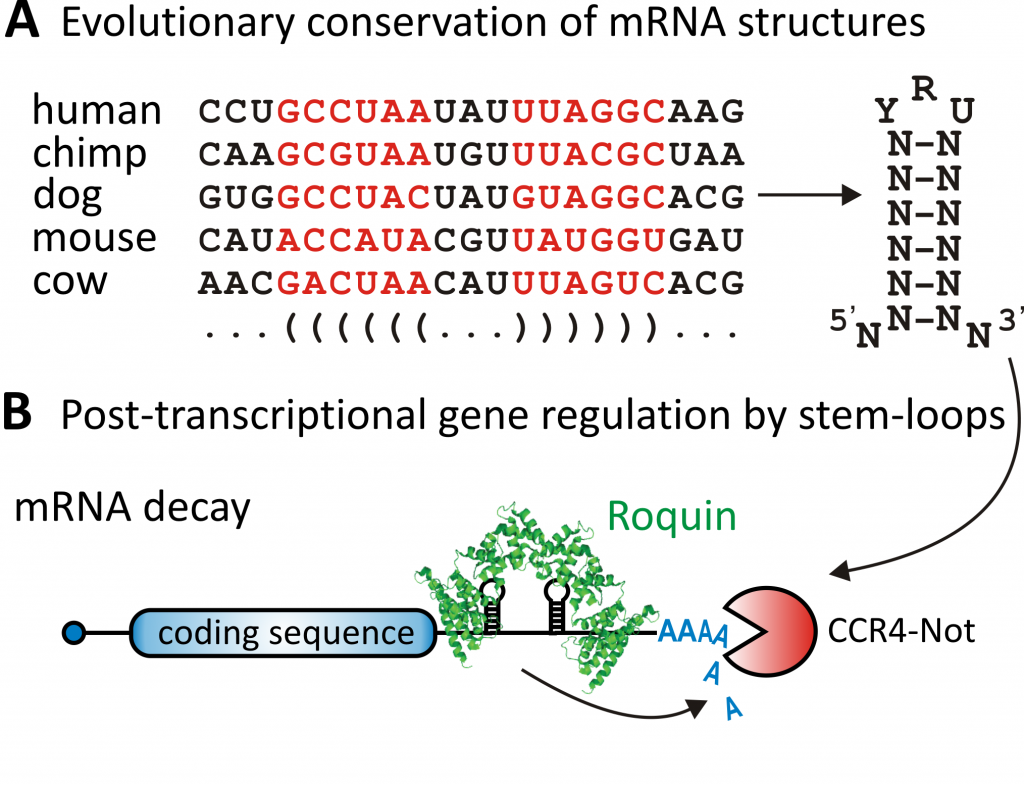
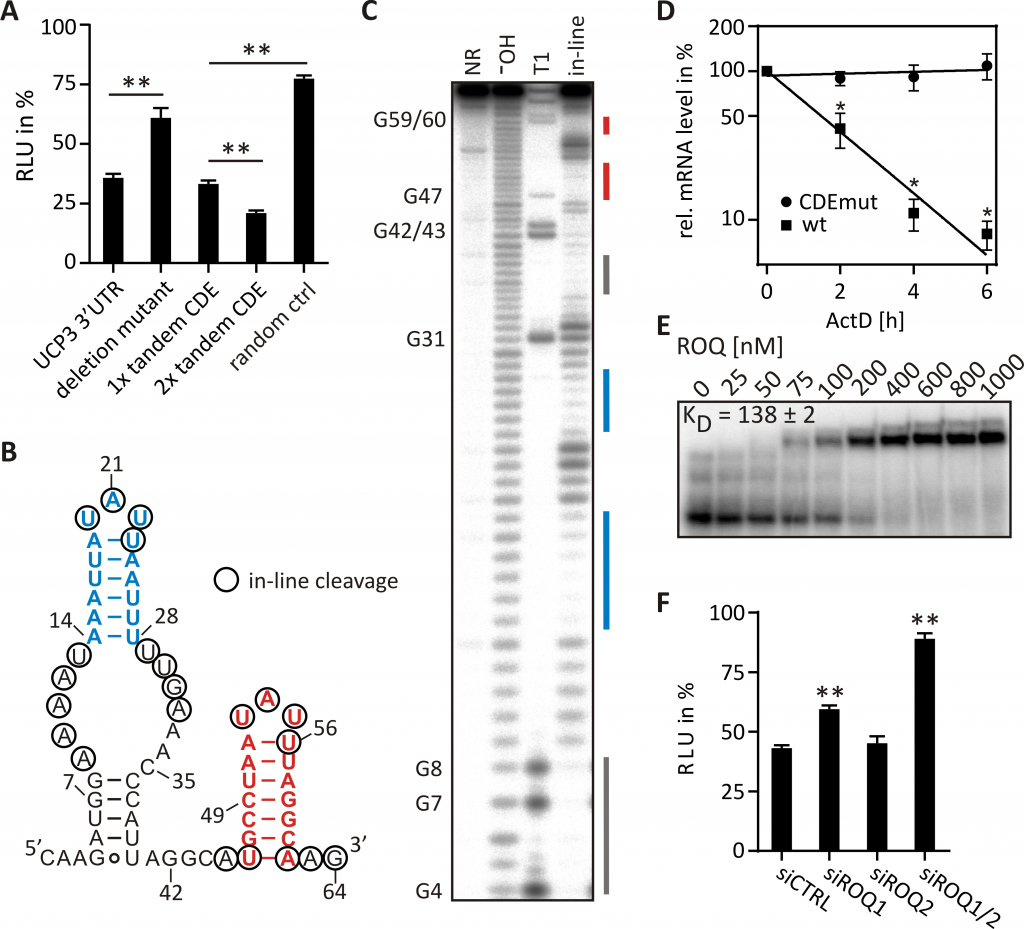
For example, we discovered a tandem CDE in the 3′-UTR of the UCP3 mRNA (Fig. 2). Structural probing and mutational analysis confirmed the importance of the two stem-loops for mRNA destabilization. Roquin binding was confirmed in vitro and post-transcriptional regulation in vivo. In-depth mutational analysis of the tandem CDE allowed us to refine the binding preferences of Roquin and to map CDEs in previously unknown target genes genome-wide. Further, we investigated the interaction of Roquin with other RNA-binding proteins that compete for mRNA binding. We discovered that AU-rich CDEs are recognized by Roquin as stem-loop and by the competitor AUF1 in their linear form, suggesting a dual function in mRNA decay depending on their folding status.
In current research, we aim to build a complete picture of Roquin binding preferences and the hierarchy of addressed target genes. Further, we are analyzing the function of other RNA structures in post-transcriptional gene regulation by massively parallel reporter gene assays and SHAPE-Map.
