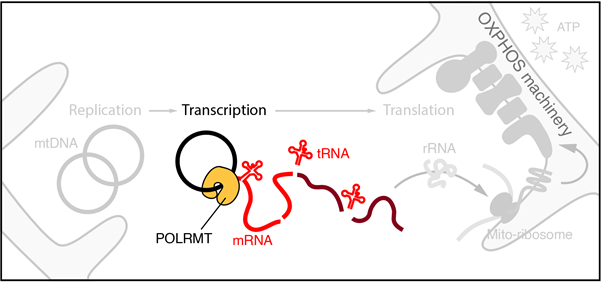
The group focuses on understanding how gene expression is regulated in mammalian mitochondria – these are organelles that form dynamic reticular networks within eukaryotic cells and are best known for their role in energy metabolism, synthesising majority of cellular adenosine triphosphate (ATP) via oxidative phosphorylation. Mitochondria are also important signalling hubs, coordinating programmed cell death (apoptosis), stress signalling, and innate immune responses.
In addition to their roles in cellular metabolism and signalling, the mitochondrial network in a mammalian cell typically contains several thousand copies of a circular, double-stranded DNA genome (mtDNA) – the genetic legacy of an ancient endosymbiotic relationship. This ‘second genome’ encodes unique protein components of the oxidative phosphorylation machinery and is indispensable for mitochondrial function and cell survival. Defects in how mtDNA is copied and expressed are heavily implicated in the ageing process, as well as in several devastating and often inherited human pathologies broadly referred to as mitochondrial disease. Currently, no known treatment exists for mitochondrial disease, and a contributing factor to this poor prognosis is an equally poor fundamental understanding of how the mitochondrial gene expression is regulated in living cells both at a basal level, and between cell types with different energetic requirements.
Our work primarily uses biochemical approaches involving purified recombinant proteins and nucleic acid substrates to reconstitute key molecular events in vitro, combining this with genetic approaches in cultured cell lines to establish the in vivo context of our work. The long-term goal of the group is to understand how mitochondrial gene expression is regulated at the level of nascent transcription, and how mitochondrial RNA metabolism gives rise to sub-mitochondrial structures known as RNA granules. Through this, we hope to better understand the pathogenesis of mitochondrial disease at a genetic level, and to identify mechanistic targets for therapeutic intervention.
Selected publications
Tan, B.G., Gustafsson, C.M. & Falkenberg, M. Mechanisms and regulation of human mitochondrial transcription. Nat Rev Mol Cell Biol. (2023). https://doi.org/10.1038/s41580-023-00661-4
Tan, B.G., Mutti, C.D.*, Shi, Y.*, Xie, X., Zhu, X., Silva-Pinheiro, P., Menger, K.E., Díaz-Maldonado, H., Wei, W., Nicholls, T.J., Chinnery P.F., Minczuk, M., Falkenberg, M., Gustafsson, C.M. (2022). The human mitochondrial genome contains a second light strand promoter. Mol Cell 82, 3646-3660.e9. https://doi.org/10.1016/j.molcel.2022.08.011. Open access. (*equal contributions)
Zhu, X.*, Xie, X.*, Das, H.*, Tan, B.G., Shi, Y., Al-Behadili, A., Peter, B., Motori, E., Valenzuela, S., Posse, V., Gustafsson, C.M., Hällberg, B.M., Falkenberg, M. (2022). Non-coding 7S RNA inhibits transcription via mitochondrial RNA polymerase dimerization. Cell 185, 2309-2323.e24. https://doi.org/10.1016/j.cell.2022.05.006. Open Access. (*equal contributions)
Tan, B.G., Wellesley, F.C., Savery, N.J., and Szczelkun, M.D. (2016). Length heterogeneity at conserved sequence block 2 in human mitochondrial DNA acts as a rheostat for RNA polymerase POLRMT activity. Nucleic Acids Res 44, 7817–7829. https://doi.org/10.1093/nar/gkw648. Open access.
